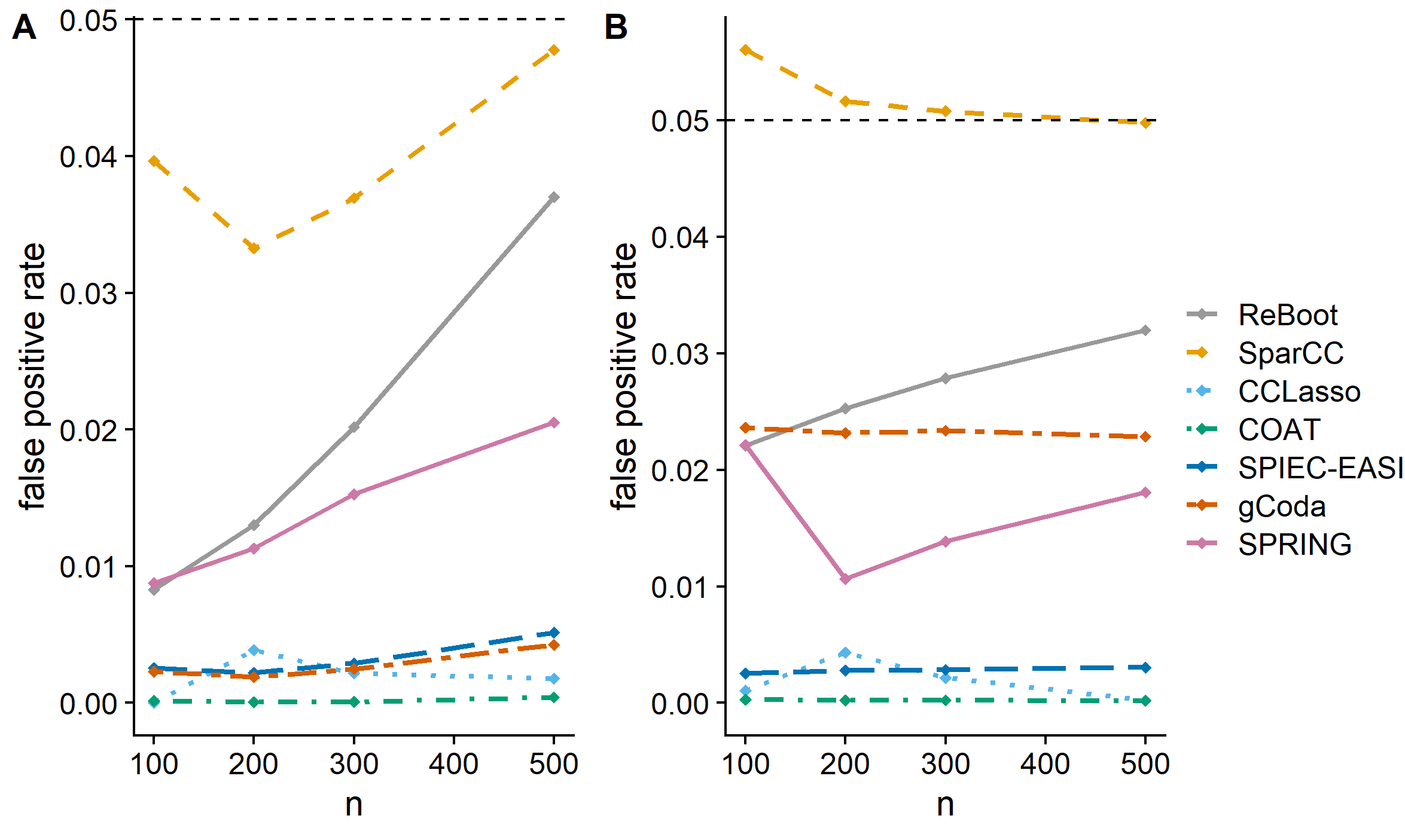Abstract
Networks are increasingly used to capture the multitude of interaction mechanisms among microbes. Due to the dynamic nature of microbial communities and their interactions, and given the paucity of well curated data bases for microbioal interactions, these networks are often inferred from microbiome abundance data. While a number of procedures have been recently proposed for this task, their properties have not been fully investigated. Two desirable properties of network estimation procedures are (i) their robustness to the spurious correlations resulting from the compositional nature of the microbiome data; and (ii) their ability to uncover the true network structure from limited observations. This chapter reviews commonly used procedures for inferring microbial interaction networks. It also presents a comprehensive empirical evaluation of these procedures.