Abstract
Motivation
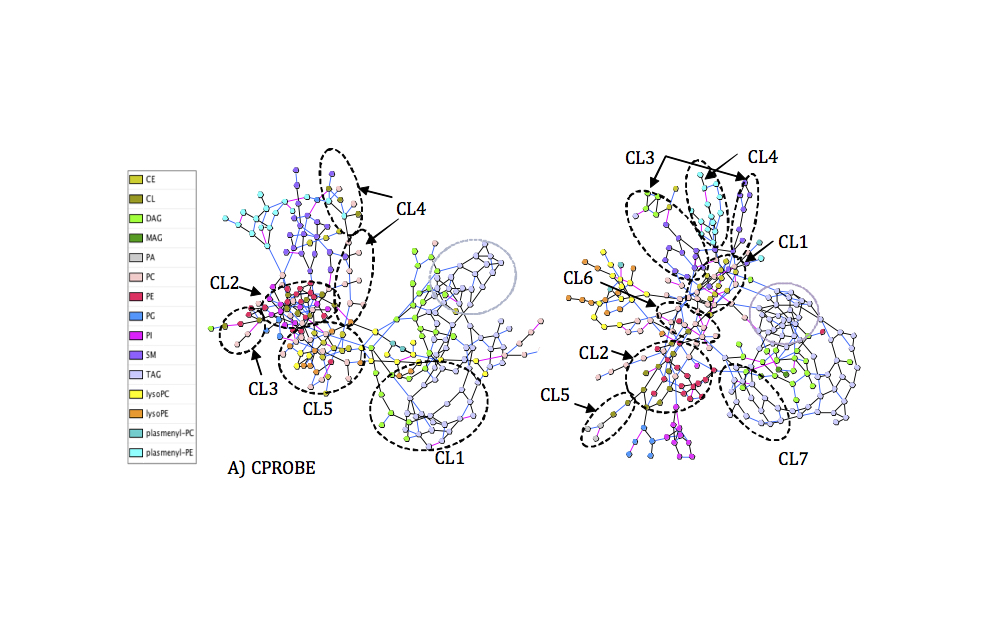
Functional enrichment testing methods can reduce data comprising hundreds of altered biomolecules to smaller sets of altered biological ‘concepts’ that help generate testable hypotheses. This study leveraged differential-network-enrichment-analysis methodology to identify and validate lipid subnetworks that potentially differentiate chronic kidney disease (CKD) by severity or progression.
Results
We built a partial-correlation interaction network, identified highly connected network components, applied network-based gene-set analysis to identify differentially enriched subnetworks, and compared the subnetworks in patients with early-stage versus late-stage CKD. We identified two subnetworks ‘triacylglycerols’ and ‘cardiolipins-phosphatidylethanolamines (CL-PE)’ characterized by lower connectivity, and a higher abundance of longer polyunsaturated triacylglycerols in patients with severe CKD (stage ≥4) from the Clinical Phenotyping Resource and Biobank Core (CPROBE). These finding were replicated in an independent cohort, the Chronic Renal Insufficiency Cohort (CRIC). Using an innovative method for elucidating biological alterations in lipid networks, we demonstrated alterations in triacylglycerols and CL-PEs that precede the clinical outcome of end-stage kidney disease by several years.
Availability
The DNEA is freely available at https://github.com/wiggie/DNEA. Java wrapper leveraging the cytoscape.js framework is available at http://js.cytoscape.org.